Learning biologically-interpretable latent representations for gene expression data
Pathway Activity Score Learning Algorithm (PASL)
Ioulia Karagiannaki, Institute of Electronic Structure and Laser, Foundation for Research and Technology-Hellas (IESL-FORTH), Heraklion; Krystallia Gourlia, Department of Computer Science, University of Crete, Heraklion; Vincenzo Lagani, JADBio Gnosis DA, Science and Technology Park of Crete, Institute of Chemical Biology, Ilia State University, Tbilisi; Yannis Pantazis, Institute of Applied and Computational Mathematics, Foundation for Research and Technology – Hellas, Heraklion; Ioannis Tsamardinos, JADBio Gnosis DA, Science and Technology Park of Crete, Department of Computer Science, University of Crete, Institute of Applied and Computational Mathematics, Foundation for Research and Technology-Hellas
Digital Library: https://link.springer.com/article/10.1007/s10994-022-06158-z
Abstract
Molecular gene-expression datasets consist of samples with tens of thousands of measured quantities (i.e., high dimensional data). However, lower-dimensional representations that retain the useful biological information do exist. A novel algorithm for such dimensionality reduction called Pathway Activity Score Learning (PASL) is presented in this paper. The major novelty of PASL is that the constructed features directly correspond to known molecular pathways (genesets in general) and can be interpreted as pathway activity scores. Hence, unlike PCA and similar methods, PASL’s latent space has a fairly straightforward biological interpretation.
Methods: PASL is shown to outperform in predictive performance the state-of-the-art method (PLIER) on two collections of breast cancer and leukemia gene expression datasets. PASL is also trained on a large corpus of 50000 gene expression samples to construct a universal dictionary of features across different tissues and pathologies. The dictionary validated on 35643 held-out samples for reconstruction error. It is then applied on 165 held-out datasets spanning a diverse range of diseases. The AutoML tool JADBio is employed to show that the predictive information in the PASL-created feature space is retained after the transformation. The code is available at https://github.com/mensxmachina/PASL.
Results
Overall, the results show that PASL (i) enables compression of the gene expression datasets that lead to 1 order of magnitude speed up in modelling, (ii) maintains the predictive information across pathologies, tissues, outcomes, and phenotypes while often leading to simpler models that are easier to interpret biologically, and (iii) complements standard GSEA in identifying differentially affected genesets across two conditions.
Conclusions: The novel PASL algorithm can help in transitioning gene expression data analysis techniques from a purely gene-centric perspective to a more systemic, pathway-centric approach.
How was JADBio used?
PASL’s ability to represent data in a latent feature space was eveluated in three aspects: (a) The out-of-sample percentage of explained variance (i.e., one minus the relative reconstruction error), (b) the predictive performance maintained for an outcome of interest in held out datasets and (c) since a PASL-constructed feature directly corresponds to known geneset, it is considered as a geneset activity score thus differential activation analysis (DAA) can be performed.
To measure the predictive performance (b) in a gene expression dataset, the automated machine learning (AutoML) tool JADBio (Tsamardinos et al., 2020) was used. The predictive performance achieved by JADBio on the original gene expression data was compared against the performance achieved by models trained on the transformed data. To ensure that a high-quality predictive model is built, JADBio searches thousands of machine learning pipelines (called configurations) to identify the optimally predictive ones and estimates the out-of-sample predictive performance of the final model in a conservative fashion.
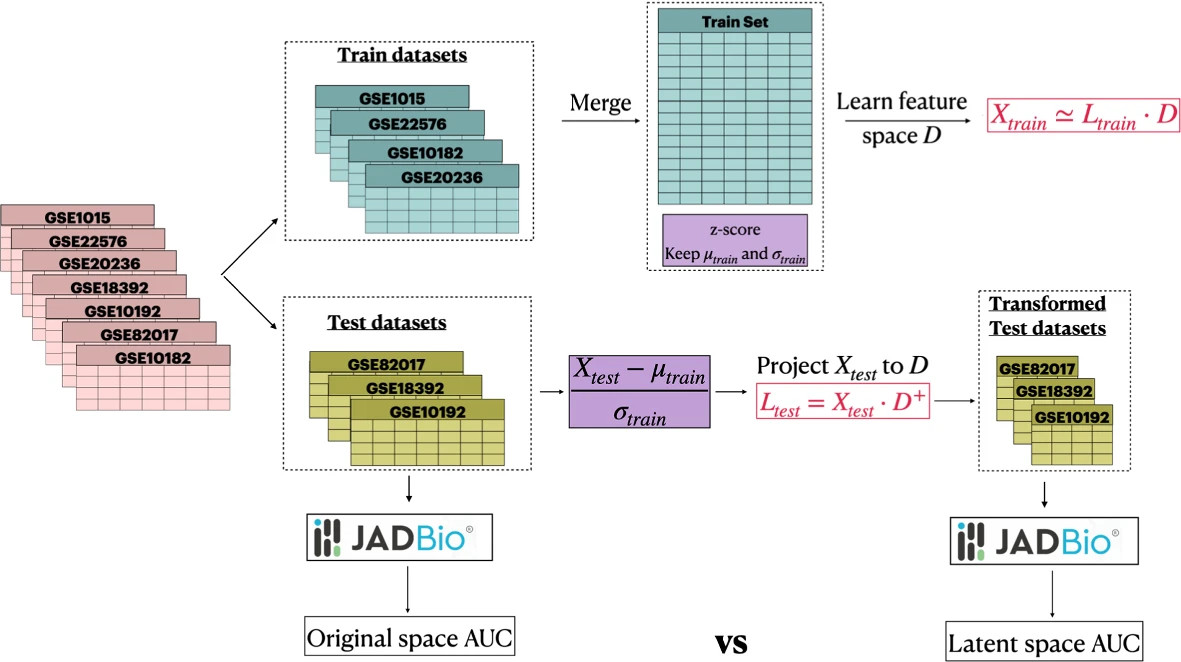
Evaluation protocol. The datasets are split into train and test datasets. The train datasets are merged creating a large dataset. PASL is applied on the train set and the final evaluation in terms of predictive performance is performed on new test datasets. The initial test datasets are compared against the lower-dimensional transformed datasets in terms of predictive performance
Illustrative case-studies of predictive modeling with JADBio in PASL space
Out of the many test datasets that were analysed by JADBio, two of them are presented, illustrating the potential advantages of predictive modeling in PASL space: a) Dataset GSE21094 containing acute Lymphoblastic Leukemia (ALL) patient data with and without Down Syndrome (Figure 10) and b) Dataset GSE30674 containing immortalized cell line data of human T lymphocyte cells (Jurkat T cells, Figure 11).
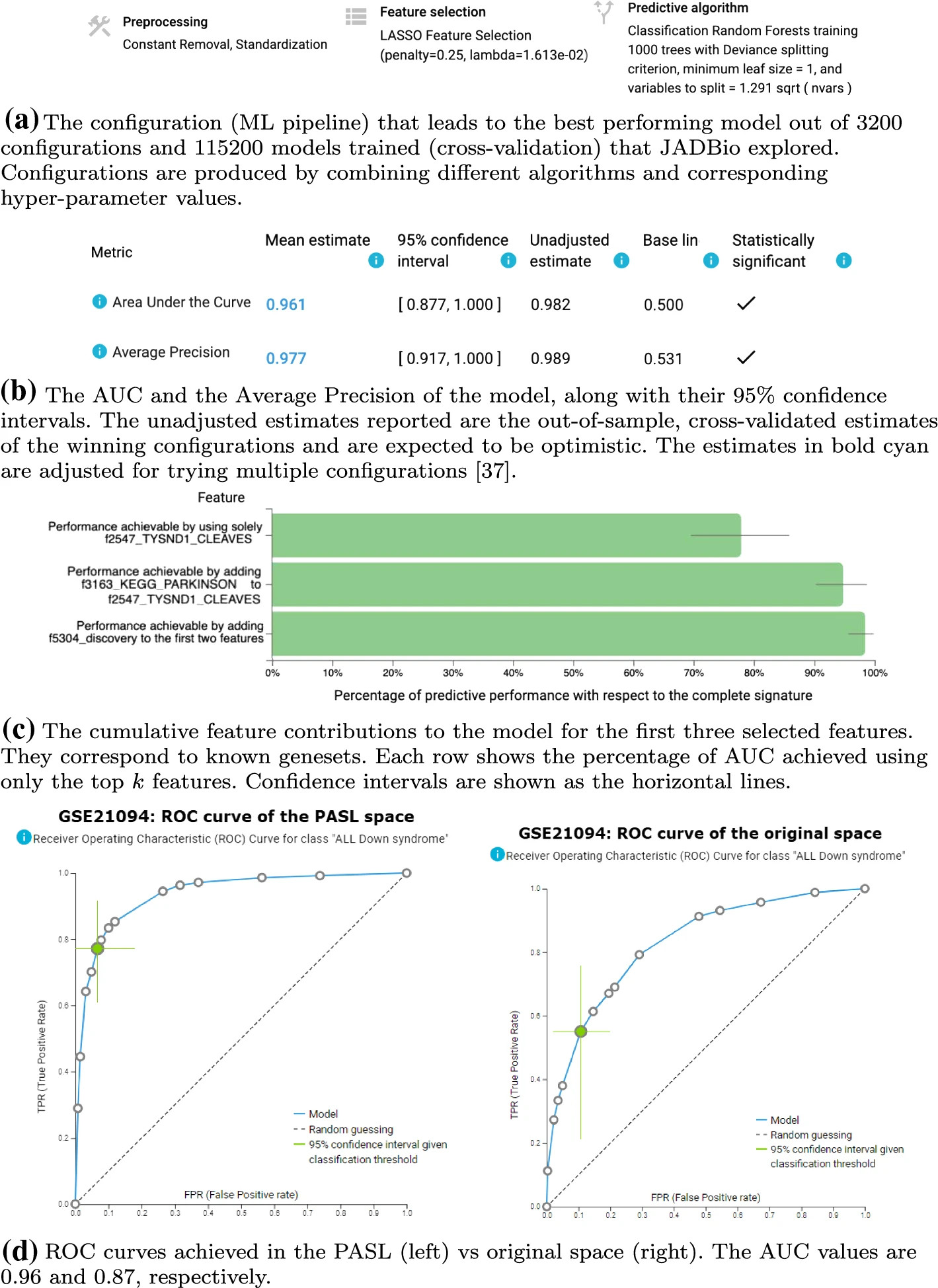
Predictive modeling with JADBio of Down Syndrome vs Non Down Syndrome in ALL cases (dataset GSE21094). Panels a, b and c (left) were produced in the PASL space (https://app.jadbio.com/share/81be792e-3ba8-43e0-ace0-b8a4fe9657f0), panel c (right) in the original space (https://app.jadbio.com/share/32892dc1-739d-4b37-b6b5-50802587fb1b)

Predictive modeling with JADBio of of kinase inhibitor treatment vs treatment control of stimuli activated Jurkat T cells (dataset GSE30674). Panels a, b and c (left) were produced in the PASL space (https://app.jadbio.com/share/9463e28b-736c-4476-8d5f-b92d7364c6aa), panel c (right) in the original space (https://app.jadbio.com/share/fe02e291-7d35-4286-bdbe-2e600b1412a3)
Read more at Machine Learning, Springer: Learning biologically-interpretable latent representations for gene expression data – PASL Algorithm
OTHER
Do you have questions?
JADBio can meet your needs. Ask one of our experts for an interactive demo.
Stay connected to get our news first!
Do you have questions?
JADBio can meet your needs. Ask one of our experts for an interactive demo.
Join the JADai Community!
Sign up with a FREE Basic plan! Be part of a growing community of AutoML enthusiasts
GET STARTED



